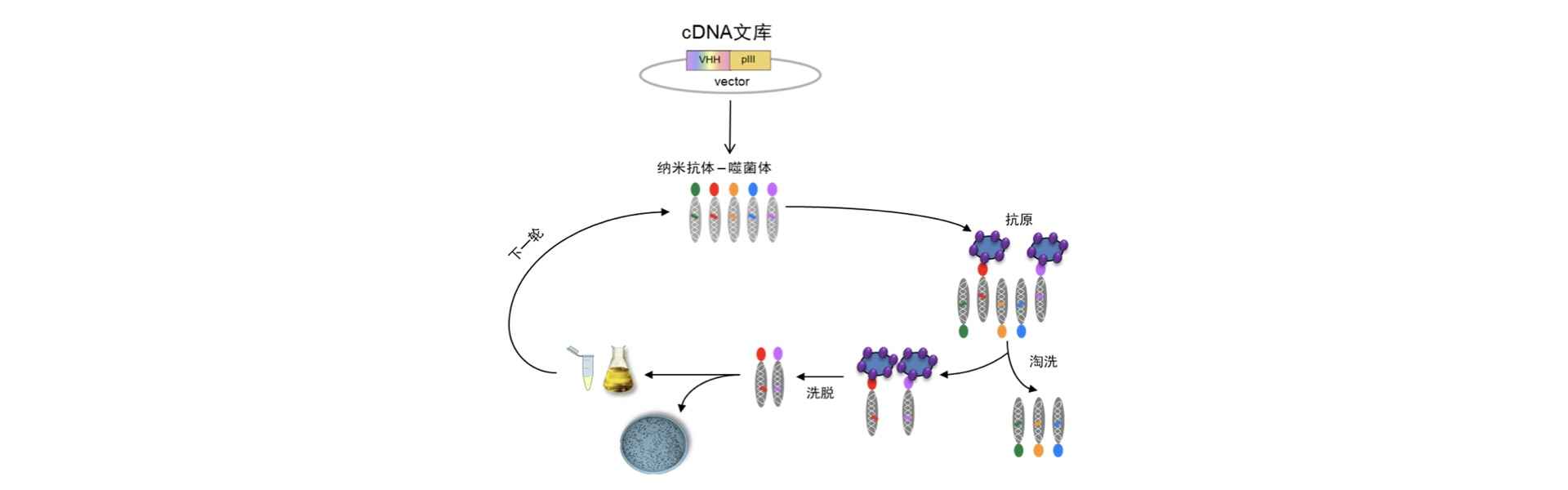
The specific steps are as follows:
- Take an appropriate amount of frozen nanobody library, inoculate into bacterial culture medium, and after suitable cultivation, add a proper amount of helper phage under suitable conditions for further cultivation.
- Extract bacteriophages amplified from the bacterial culture supernatant using the PEG-NaC method.
- Incubate the bacteriophages with the antigen. If the antigen is a cell membrane protein, incubate bacteriophages with whole cells or cell membrane extracts enriched with overexpressed antigens. If the antigen is intracellular or secreted, pre-fix the antigen on a medium such as a test tube or microplate and then incubate with bacteriophages.
- Wash the mixture. Discard bacteriophages and wash the antigen several times with an appropriate buffer (such as PBS) to remove non-specifically bound bacteriophages, retaining those specifically bound to the antigen.
- Elute. Treat bacteriophages specifically bound to the antigen with an appropriate method (such as acidic glycine solution) to dissociate bacteriophages from the antigen and preserve them.
At this point, bacteriophages expressing specific nanobodies are obtained. These bacteriophages can be used for the following technical operations:
- Transformation into a specific nanobody library: Infect bacteriophages into Escherichia coli at an appropriate state without adding helper phages. Once bacteriophage infection is complete, specific nanobodies exist in Escherichia coli in the form of DNA plasmids. Collect these Escherichia coli, and it becomes a nanobody library specific to the antigen. This library can be used as raw material to return to step (1) for the next round of bacteriophage surface display screening.
- Transformation into monoclonal nanobody colonies: Take a small amount of bacteriophages obtained in step (5), dilute them, and then infect Escherichia coli at a suitable state without adding helper phages. Once bacteriophage infection is complete, evenly spread these Escherichia coli on bacterial culture dishes, adjust the culture appropriately to obtain monoclonal colonies containing nanobody DNA plasmids. Use these monoclonal colonies as raw material for positive monoclonal nanobody identification.
Note: The translation provided is technical and may require further clarification based on the specific context and terminology used in the original text.
Upon obtaining bacterial culture dishes with colonies, positive monoclonal nanobodies can be identified. The specific technical scheme is as follows:
- Pick a single colony and cultivate it in a microplate.
- Induce the expression of VHH-pIII (the fusion protein containing nanobody) by adding IPTG.
- Collect the bacterial culture supernatant containing nanobodies and incubate it with the antigen.
- Use methods such as flow cytometry to detect whether the monoclonal nanobodies bind to the antigen.
- For microbial colonies with monoclonal nanobodies that can bind to the antigen, after appropriate cultivation, extract plasmid DNA and perform DNA sequencing to obtain the gene sequence of the nanobody. Upon translation, the complete amino acid sequence of the nanobody can be obtained.
Note: The translation provided is technical and may require further clarification based on the specific context and terminology used in the original text.