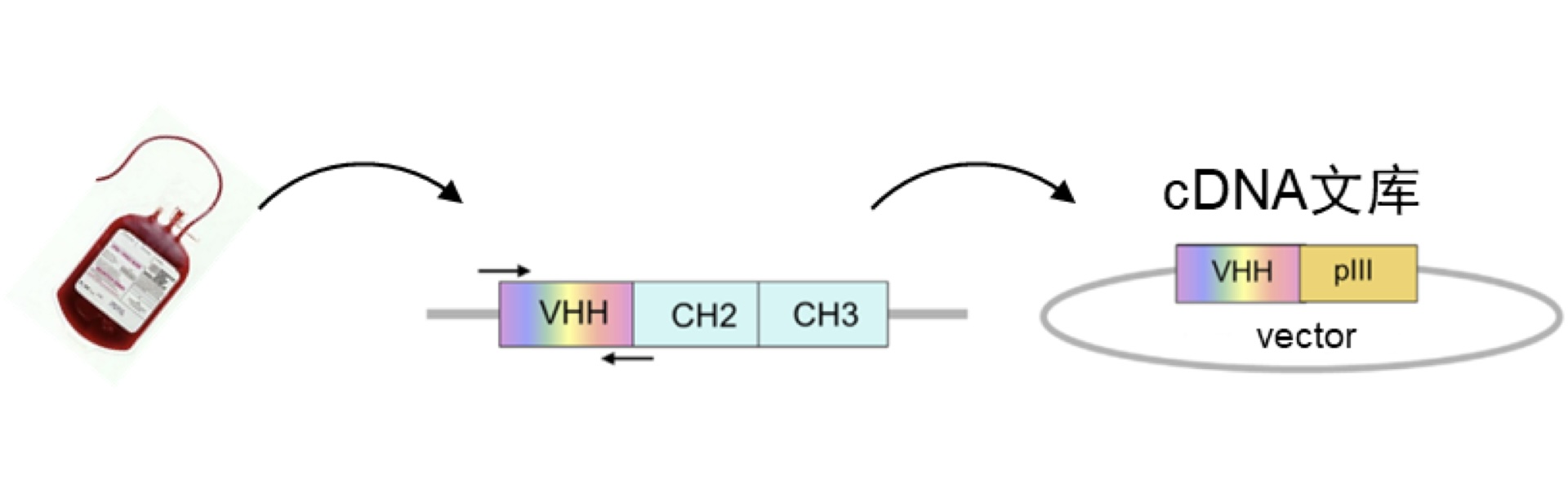
Constructing a High-Diversity Nanobody DNA Library involves a two-phase blood collection process. The first round of blood collection occurs after the completion of the third antigen immunization, and the second round follows the fourth antigen immunization, with a total blood volume exceeding 200mL. Lymphocytes are isolated, and RNA extraction, along with the construction of the library, is performed.
The library construction plan is as follows:
-
Isolate lymphocytes from alpaca peripheral blood using methods such as density gradient centrifugation.
-
Extract total mRNA from the isolated lymphocytes and reverse transcribe it into cDNA.
-
Use appropriate DNA primers to amplify the VHH fragments of alpaca immunoglobulins IgG2 and IgG3 through polymerase chain reaction (PCR) using the aforementioned cDNA as a template. These VHH fragments represent the DNA segments of nanobodies.
-
Connect the DNA fragments of VHH to a phage display screening vector on the surface of bacteriophage, forming a VHH-pIII fusion protein expression vector plasmid library. Here, pIII is a protein present on the surface flagella of the bacteriophage.
-
Utilize electroporation to transform the DNA construct into TG1 competent bacteria.
-
Under suitable culture conditions, collect all bacterial colonies, resulting in the alpaca nanobody library
Compared to traditional methods of isolating antibodies from the serum or lymphocytes of mice, rabbits, and other animals, this technological approach allows for the long-term preservation of the complete nanobody fragments obtained from alpacas (i.e., the library). This capability ensures continuous support for ongoing nanobody screening and development. The library's diversity is maintained at 10^8-10^9 colony-forming units (cfu).