Now, without the need for alpaca immunization, desired nanobody sequences can be obtained through screening of natural libraries. The alpaca VHH natural library constructed by Nanobo Life Sciences contains a diverse range of VHH gene sequences from different alpaca individuals, allowing for direct screening upon customer antigen provision. The advantages of natural libraries include short cycles, large capacity, and high quality. With a capacity of 1.3*10^11 cfu/mL and high-quality output, specific nanobody sequences can be obtained in 2-3 weeks. This approach is suitable for toxic antigens to alpacas as well as antigens with low immunogenicity.

Construction and process testing of the alpaca VHH natural library
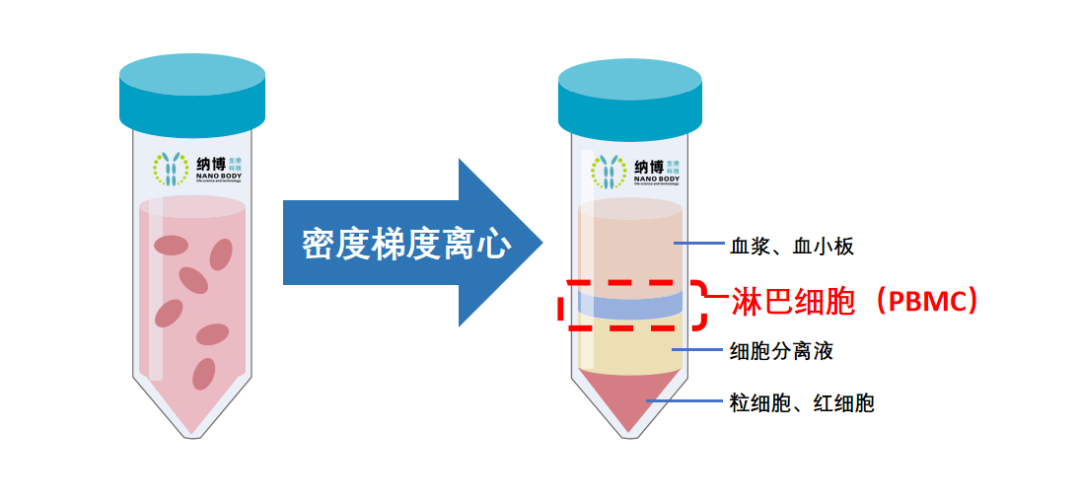
The construction of the alpaca VHH natural library begins with the isolation of peripheral blood mononuclear cells (PBMCs). Below are the steps and procedures:
-
Add an equal volume of D-PBS diluent to venous peripheral blood containing anticoagulant, and mix well.
-
Inject lymphocyte separation solution into density gradient centrifuge tubes, adding 15 mL per tube.
-
Keep the density gradient centrifuge tube vertical, and slowly add the diluted venous peripheral blood along the tube wall using a serological pipette. The PBMC sample will mix with the separation solution above the partition, but this does not affect the separation effect.
-
Using a horizontal rotor centrifuge, centrifuge at 1200g for 10 minutes at room temperature with the centrifuge motor activated.
-
After centrifugation, remove the cell-free plasma and D-PBS diluent from the top layer of the density gradient centrifuge tube, retaining 5 mL of plasma for titer testing.
-
Slowly aspirate the upper layer diluent containing lymphocytes into new sterile centrifuge tubes, add an equal volume of D-PBS, and centrifuge at 1000g for 10 minutes at room temperature to wash the cells.
-
After centrifugation, discard the supernatant, and freeze the PBMCs obtained from the bottom sediment at -80°C in an ultra-low temperature freezer.
Phage display library construction process
Phage display library construction process includes: total RNA extraction, reverse transcription, PCR amplification, enzyme digestion and ligation, electroporation into TG1 competent cells, and finally, library collection.
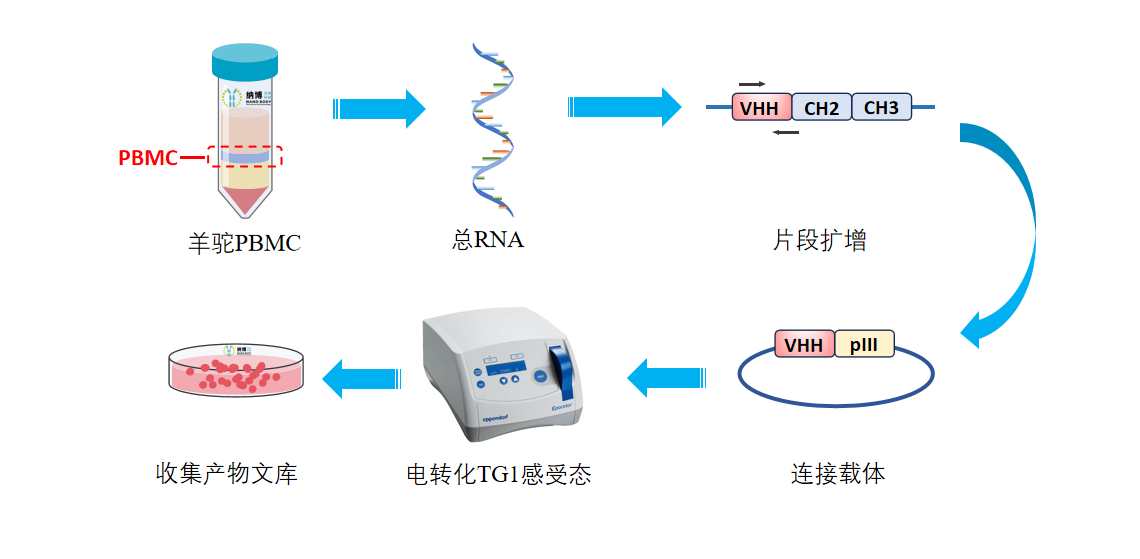
The diagram illustrates the schematic representation of the phage display library construction process.
Construction of Phage Display Library—Experimental Procedure Overview
(1) Total RNA Extraction: Merge all extracted PBMCs and use column adsorption to extract total mRNA.
(2) Reverse Transcription: Utilize the extracted total mRNA as a template for reverse transcription to obtain cDNA.
(3) PCR: Amplify the VHH fragments of alpaca immunoglobulins IgG2 and IgG3, i.e., the DNA fragments of nanobodies, using appropriate DNA primers and the aforementioned cDNA template through polymerase chain reaction (PCR).
(4) Enzyme Digestion and Ligation: (a) Vector Enzyme Digestion: Linearize the phage display selection vector DNA plasmid through double enzyme digestion. (b) Fragment Enzyme Digestion: Digest the VHH DNA fragments with two enzymes. (c) Vector and Fragment Ligation: Employ T4 DNA ligase to ligate the digested vector and fragments, resulting in the complete VHH phage display selection plasmid library, i.e., VHH-pIII fusion protein expression vector plasmid library. Here, pIII is a protein present on the flagella of the phage.
(5) Electroporation into TG1 Competent Cells: Utilize electroporation to transform the DNA ligation products into TG1 Escherichia coli competent cells, with 96 transformations conducted.
(6) Library Collection: Cultivate all collected colonies appropriately, constituting the nanobody library of immunized alpacas. Throughout this process, the size of the library before cultivation is measured using gradient dilution, estimated to be approximately 1.56×10^9 cfu.
(1) Randomly select no fewer than 48 microbial monoclonal clones from the library. After overnight incubation at 37°C, extract plasmid DNA and perform Sanger sequencing. (2) Translate all sequencing results to obtain 48 correct and valid nanobody sequences (as shown in the attachment), including 48 completely different CDR3 regions (as illustrated in the figure).

Figure: Alignment of the amino acid sequences of the CDR3 regions from 48 sequencing results"
Fragment Insertion Rate Testing
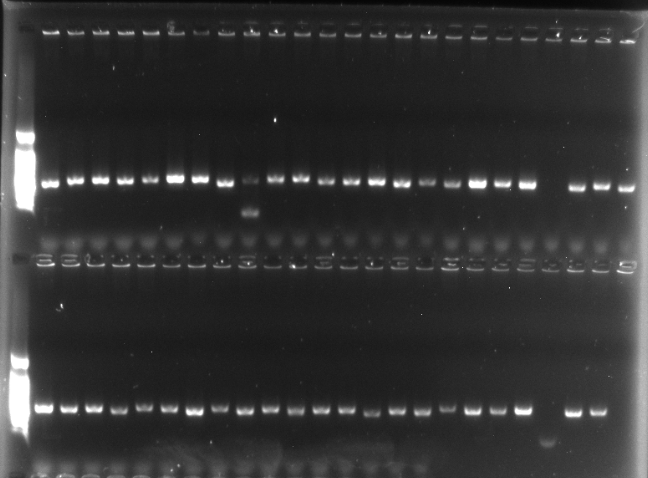
(1) Perform PCR amplification on the VHH fragments from the aforementioned 48 microbial monoclonal clones. (2) Analyze the PCR products by agarose gel electrophoresis to evaluate the insertion rate of the VHH fragments.
Figure: Agarose Gel Electrophoresis Analysis of PCR Products from 48 VHH Gene Fragments
From the results of the gel electrophoresis analysis, it is observed that out of the 48 microbial monoclonal clones, 45 clones successfully amplified the VHH gene fragments via PCR, while the remaining 3 clones failed to amplify. Considering that the Sanger sequencing of the VHH fragments was all correct as mentioned earlier, the inability to amplify the VHH fragments in these 3 clones is attributed to experimental error rather than sequence diversity. Thus, it does not affect the assessment of the diversity of the samples.