-
Antigen Preparation and Alpaca Immunization
Target:
GLP1R - Glucagon-like peptide 1 receptor
Using HEK293 cell line stably expressing GLP1R as the antigen.
Antigen form: Cell membrane extract stably expressing GLP1R
Cell quantity: 3*10^8 cells
Number of immunizations: Four immunizations
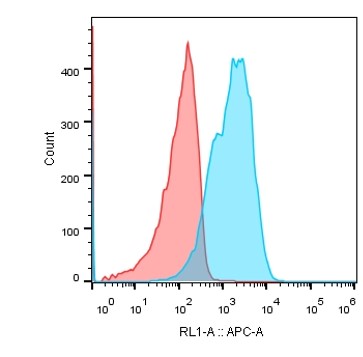
Stable cell line expressing GLP1R
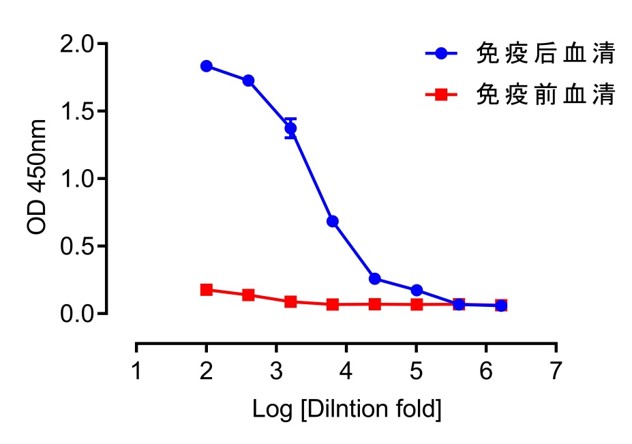
| Dilution ratio | Post-immunization serum | Pre-immunization serum | ||||
| 100 | 1.829 | 1.833 | 1.841 | 0.211 | 0.158 | 0.162 |
| 400 | 1.723 | 1.75 | 1.705 | 0.147 | 0.135 | 0.134 |
| 1600 | 1.455 | 1.329 | 1.337 | 0.088 | 0.092 | 0.085 |
| 6400 | 0.722 | 0.659 | 0.671 | 0.064 | 0.07 | 0.066 |
| 25600 | 0.297 | 0.247 | 0.233 | 0.066 | 0.075 | 0.066 |
| 102400 | 0.15 | 0.18 | 0.19 | 0.065 | 0.068 | 0.069 |
| 409600 | 0.055 | 0.068 | 0.079 | 0.074 | 0.071 | 0.064 |
| 1638400 | 0.055 | 0.061 | 0.058 | 0.057 | 0.064 | 0.065 |
-
Phage Library Construction
Total library size after electroporation: approximately 9.00×10^8 cfu. A small aliquot of the library was retained after electroporation and before amplification. From this, 48 individual clones were randomly selected after amplification for the following experiments:
PCR was performed to assess the insertion rate of the phage vector fragments. Using specific primers, the VHH gene fragments of 48 individual clones were amplified. Gel electrophoresis revealed that 4 clones did not correctly insert the VHH gene fragment.
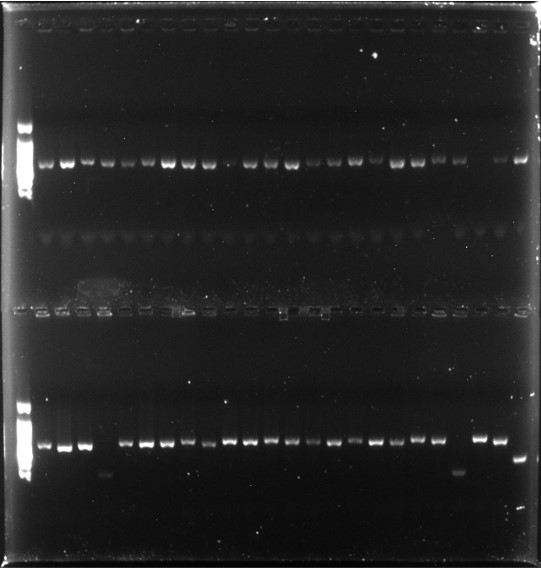
Insertion rate = (44/48) × 100% = 91.67%
Sanger sequencing was employed to assess the sequence diversity.
After obtaining the DNA sequences, they were translated into amino acid sequences. Sequences that failed, were invalid, or were duplicates were removed. Subsequently, multiple sequence alignment was conducted using MAFFT, resulting in 38 unique VHH fragments with completely different CDR3 regions.

Library CDR3 diversity = (9.00×10^8) × (38/48) = 7.125×10^8 cfu
3. Phage Surface Display Screening
First Round Preliminary Screening: A total of 4 96-well plates of microbial clones were selected for flow cytometry analysis screening, including a large number of non-specific clones. Detailed experimental data can be found in the following page.
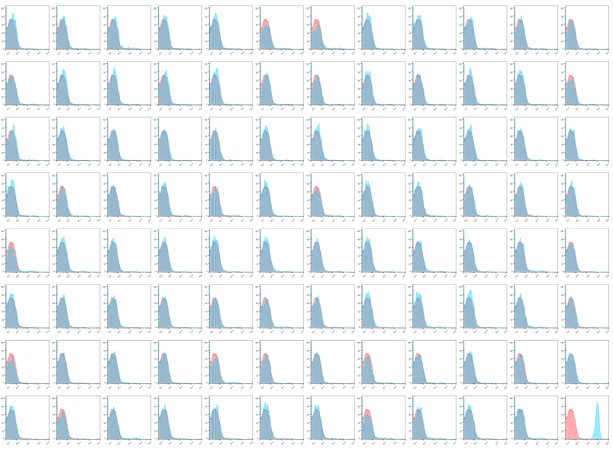
Results of the First Round Screening Flow Cytometry Analysis
Second Round Preliminary Screening: A total of 4 96-well plates of microbial clones were selected for flow cytometry analysis screening, including a large number of positive clones. Detailed experimental data can be found in the following page.
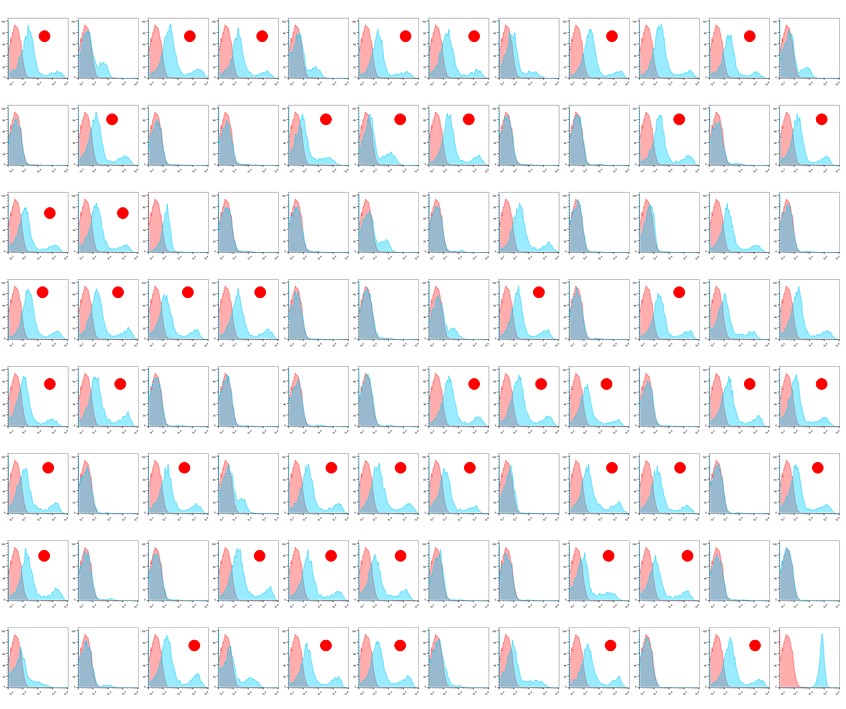
Results of the Second Round Screening Flow Cytometry Analysis
Conclusion: The first round yielded no positive clones, while the second round screening resulted in nearly 300 positive clones
Note: Detection was performed using HEK293T cells transiently expressing GLP1R. Flow cytometry analysis revealed an irregular bimodal distribution of positive results. Results marked with red dots indicate preliminary positive outcomes.
4.Sequencing revealed positive VHH sequences.
- After sequencing, a total of 260 valid VHH gene sequences were obtained from the approximately 300 clones.
- Following translation and filtering out errors and duplicates, 52 unique VHH gene sequences remained.
- These 52 clones were then recombinantly expressed and purified.
- Flow cytometry analysis confirmed the affinity between the nanobodies and GLP1R, resulting in the identification of 47 effective nanobodies.
- These nanobodies were further evaluated for their binding to GLP1R using HEK293T cells stably expressing GLP1R.
5. The flow cytometry results of monoclonal nanobodies binding to GLP-1R are as follows:
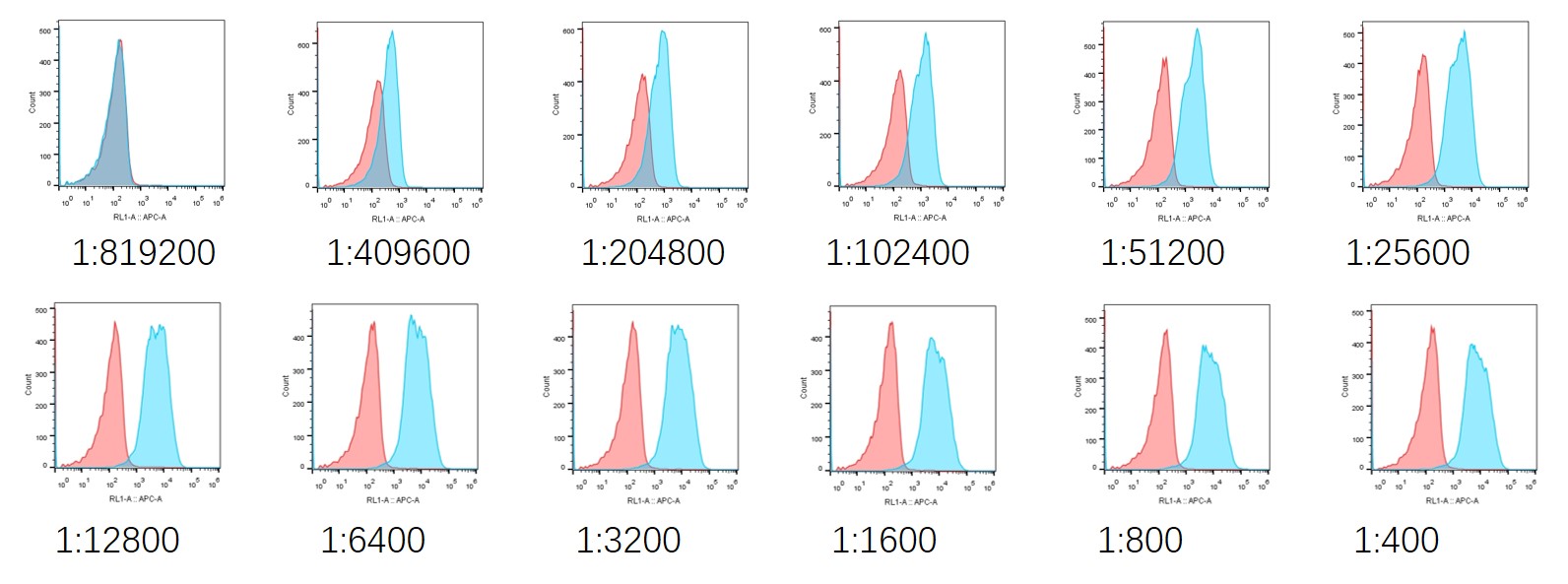
The results of the detection at different dilution ratios are as follows:
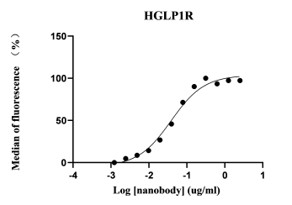
The initial dilution was performed at 1:400 (2.5ug/ml), and then further diluted by a factor of 2.