This case study consists of four steps: immunization of camels, construction of phage display libraries, phage surface display screening, and Sanger sequencing to obtain positive VHH sequences.
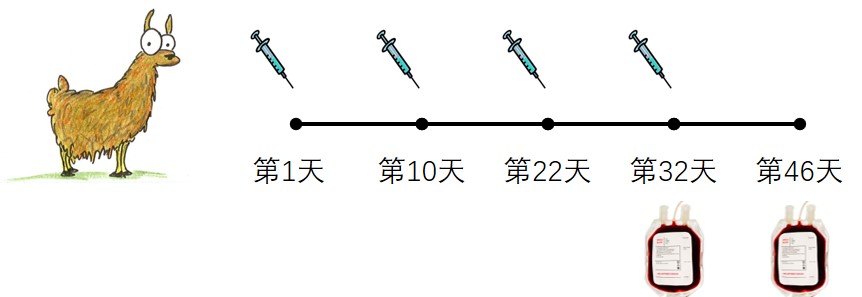
1- AlpacaImmunization: Immunization of Alpacas with purified IL-18 protein
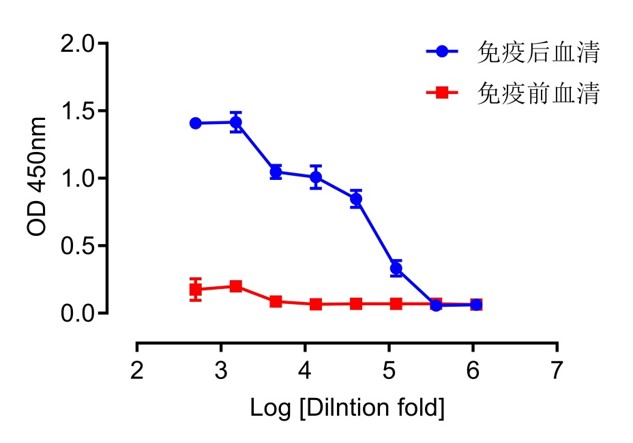
| Dilution Ratio | Post-Immune Serum | Pre-Immune Serum | ||||
| 500 | 1.429 | 1.43 | 1.367 | 0.263 | 0.156 | 0.105 |
| 1500 | 1.333 | 1.474 | 1.438 | 0.169 | 0.204 | 0.224 |
| 4500 | 1.046 | 1.097 | 1.001 | 0.095 | 0.076 | 0.087 |
| 13500 | 0.963 | 0.956 | 1.105 | 0.068 | 0.068 | 0.061 |
| 40500 | 0.893 | 0.775 | 0.874 | 0.067 | 0.067 | 0.072 |
| 121500 | 0.361 | 0.265 | 0.369 | 0.066 | 0.068 | 0.073 |
| 364500 | 0.061 | 0.05 | 0.06 | 0.075 | 0.075 | 0.06 |
| 1093500 | 0.059 | 0.064 | 0.065 | 0.054 | 0.069 | 0.070 |
2 - Construction of Phage Display Library
Total Library Size after Electroporation:
Approximately 7.30×10^8 cfu. Following electroporation and before amplification, a small aliquot of the sample was taken. After amplification culture, 48 individual clones were randomly selected for the following experiments:
PCR Evaluation of Phage Vector Fragment Insertion Rate:
Using specific primers, PCR amplification of the VHH gene fragments from 48 individual clones was performed. Agarose gel electrophoresis revealed that four clones did not contain the correct insertion of the VHH gene fragment.
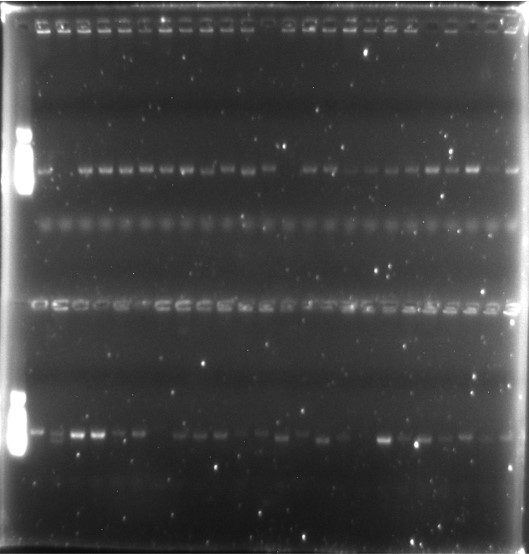
Fragment Insertion Rate: 4448×100%=91.67%
Sanger Sequencing to Assess Sequence Diversity
After obtaining the DNA sequences, they were translated into amino acid sequences. Sequences that failed sequencing, were invalid, or were duplicated were removed. Using MAFFT for multiple sequence alignment, a total of 38 unique VHH fragments with completely different CDR3 regions were obtained (see the figure below).

Library CDR3 Diversity: 7.30×108×38/48=5.78×108 cfu
3 - Phage Surface Display Screening
First Round Initial Screening: A total of 4 microplate clones were picked for ELISA screening. Detailed experimental data are shown in the table below.
Conclusion: The first round of initial screening identified 5 potential positive clones.
plate 1
| / | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| A | 0.0707 | 0.0735 | 0.0791 | 0.0756 | 0.0751 | 0.0728 | 0.0735 | 0.074 | 0.0714 | 0.0746 | 0.0717 | 0.0799 |
| B | 0.1635 | 0.0763 | 0.075 | 0.0823 | 0.0808 | 0.0803 | 0.0746 | 0.0751 | 0.0773 | 0.0769 | 0.0706 | 0.0707 |
| C | 0.0741 | 0.0748 | 0.0747 | 0.0758 | 0.0887 | 0.0835 | 0.0766 | 0.0759 | 0.0753 | 0.0778 | 0.0735 | 0.0717 |
| D | 0.0714 | 0.0777 | 0.0791 | 0.0749 | 0.0783 | 0.0782 | 0.0773 | 0.0773 | 0.0776 | 0.0806 | 0.0759 | 0.0716 |
| E | 0.0738 | 0.0752 | 0.074 | 0.077 | 0.0743 | 0.0748 | 0.0762 | 0.0786 | 0.0758 | 0.0756 | 0.0788 | 0.0746 |
| F | 0.0795 | 0.0811 | 0.0819 | 0.0738 | 0.0827 | 0.0773 | 0.0771 | 0.0769 | 0.0731 | 0.0808 | 0.074 | 0.074 |
| G | 0.0734 | 0.0729 | 0.0732 | 0.0756 | 0.077 | 0.0743 | 0.0801 | 0.0799 | 0.0805 | 0.0808 | 0.0731 | 0.0642 |
| H | 0.0692 | 0.0729 | 0.0732 | 0.0741 | 0.0785 | 0.0793 | 0.0795 | 0.0778 | 0.0739 | 0.0731 | 0.0735 | 0.1851 |
plate 2
| / | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| A | 0.0712 | 0.0788 | 0.0778 | 0.0757 | 0.0766 | 0.0762 | 0.0822 | 0.0822 | 0.0764 | 0.077 | 0.0739 | 0.0883 |
| B | 0.0784 | 0.0754 | 0.0861 | 0.0732 | 0.0767 | 0.0921 | 0.077 | 0.0837 | 0.0773 | 0.0735 | 0.074 | 0.0786 |
| C | 0.0864 | 0.0818 | 0.0775 | 0.078 | 0.078 | 0.079 | 0.0781 | 0.0771 | 0.0825 | 0.0782 | 0.0804 | 0.0782 |
| D | 0.0784 | 0.1443 | 0.0788 | 0.0837 | 0.0801 | 0.0771 | 0.075 | 0.0814 | 0.0818 | 0.0971 | 0.0818 | 0.0761 |
| E | 0.0726 | 0.0778 | 0.0784 | 0.0843 | 0.0889 | 0.078 | 0.0781 | 0.0779 | 0.0772 | 0.0758 | 0.0757 | 0.0777 |
| F | 0.0769 | 0.079 | 0.0789 | 0.0827 | 0.0811 | 0.0783 | 0.0803 | 0.0836 | 0.0883 | 0.0846 | 0.0828 | 0.0817 |
| G | 0.0722 | 0.0792 | 0.0788 | 0.0767 | 0.0819 | 0.0839 | 0.0777 | 0.0766 | 0.0778 | 0.083 | 0.0753 | 0.0704 |
| H | 0.0735 | 0.0768 | 0.0798 | 0.0805 | 0.0756 | 0.0758 | 0.0738 | 0.0768 | 0.0807 | 0.0832 | 0.0812 | 0.189 |
plate 3
| / | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| A | 0.0694 | 0.0757 | 0.0786 | 0.0755 | 0.0804 | 0.0745 | 0.0788 | 0.0774 | 0.0825 | 0.0756 | 0.0775 | 0.0763 |
| B | 0.0702 | 0.0786 | 0.0798 | 0.0794 | 0.0814 | 0.0813 | 0.0822 | 0.0818 | 0.0814 | 0.0775 | 0.0771 | 0.076 |
| C | 0.0709 | 0.0814 | 0.0784 | 0.086 | 0.0779 | 0.0786 | 0.0799 | 0.0776 | 0.0794 | 0.0761 | 0.075 | 0.076 |
| D | 0.0757 | 0.0799 | 0.0792 | 0.0802 | 0.0862 | 0.0799 | 0.0782 | 0.0791 | 0.0785 | 0.0839 | 0.08 | 0.0769 |
| E | 0.0716 | 0.0775 | 0.0754 | 0.0774 | 0.0847 | 0.0803 | 0.083 | 0.0758 | 0.0801 | 0.0791 | 0.0764 | 0.0802 |
| F | 0.0748 | 0.0772 | 0.0791 | 0.0804 | 0.0906 | 0.08 | 0.0808 | 0.0813 | 0.0784 | 0.0764 | 0.0808 | 0.0771 |
| G | 0.0738 | 0.0787 | 0.0787 | 0.08 | 0.082 | 0.0798 | 0.0815 | 0.0785 | 0.0816 | 0.0792 | 0.0761 | 0.071 |
| H | 0.073 | 0.082 | 0.0762 | 0.1198 | 0.0831 | 0.0814 | 0.0821 | 0.0787 | 0.0804 | 0.0753 | 0.0799 | 0.2075 |
Second Round Initial Screening: A total of 4 microplate clones were picked for ELISA screening (plates 05-08, with 2 control clones on each plate). Detailed experimental data are shown in the table below.
Conclusion: The second round of initial screening identified approximately 90 potential positive clones.
plate 05
| / | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| A | 0.2861 | 0.0619 | 0.1553 | 0.0571 | 0.0697 | 0.1173 | 0.2654 | 0.0631 | 0.1353 | 0.0567 | 0.0676 | 0.0561 |
| B | 0.174 | 0.0606 | 0.0587 | 0.0583 | 0.063 | 0.1602 | 0.056 | 0.0586 | 0.0623 | 0.1347 | 0.0615 | 0.056 |
| C | 0.0613 | 0.0912 | 0.0566 | 0.2031 | 0.0758 | 0.0636 | 0.0978 | 0.0585 | 0.1514 | 0.1922 | 0.0553 | 0.0561 |
| D | 0.063 | 0.1056 | 0.098 | 0.0726 | 0.0943 | 0.0655 | 0.061 | 0.0656 | 0.0557 | 0.1594 | 0.0552 | 0.0607 |
| E | 0.0605 | 0.1982 | 0.0564 | 0.1177 | 0.0627 | 0.0672 | 0.0604 | 0.0769 | 0.0608 | 0.0617 | 0.0634 | 0.063 |
| F | 0.0596 | 0.0609 | 0.0579 | 0.0616 | 0.0588 | 0.0642 | 0.0574 | 0.058 | 0.0568 | 0.1712 | 0.0987 | 0.0556 |
| G | 0.0565 | 0.0783 | 0.0568 | 0.2033 | 0.0826 | 0.0711 | 0.0556 | 0.1341 | 0.0553 | 0.1082 | 0.0548 | 0.0559 |
| H | 0.0827 | 0.2919 | 0.091 | 0.2727 | 0.0567 | 0.1121 | 0.0568 | 0.2702 | 0.0873 | 0.255 | 0.056 | 0.251 |
plate 06
| / | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| A | 0.0651 | 0.0566 | 0.0684 | 0.102 | 0.0653 | 0.0597 | 0.2466 | 0.0888 | 0.0683 | 0.2014 | 0.0899 | 0.0562 |
| B | 0.1529 | 0.0582 | 0.2096 | 0.057 | 0.0747 | 0.062 | 0.0555 | 0.0575 | 0.0657 | 0.0554 | 0.2129 | 0.055 |
| C | 0.0559 | 0.0596 | 0.2581 | 0.0582 | 0.0591 | 0.0569 | 0.1052 | 0.0566 | 0.0508 | 0.063 | 0.1112 | 0.0552 |
| D | 0.0972 | 0.2394 | 0.0759 | 0.0588 | 0.0582 | 0.0606 | 0.2575 | 0.0711 | 0.0555 | 0.0583 | 0.0557 | 0.0571 |
| E | 0.0579 | 0.0591 | 0.0578 | 0.0659 | 0.0974 | 0.0647 | 0.0575 | 0.2389 | 0.0557 | 0.0598 | 0.0582 | 0.0616 |
| F | 0.0591 | 0.0707 | 0.1009 | 0.0634 | 0.0777 | 0.0636 | 0.0592 | 0.0594 | 0.0572 | 0.0601 | 0.0648 | 0.0566 |
| G | 0.0574 | 0.1157 | 0.0565 | 0.0588 | 0.0554 | 0.1596 | 0.1966 | 0.0673 | 0.0539 | 0.1265 | 0.064 | 0.0552 |
| H | 0.0968 | 0.0674 | 0.0932 | 0.059 | 0.0571 | 0.0747 | 0.0819 | 0.0662 | 0.123 | 0.2302 | 0.1021 | 0.2622 |
plate 07
| / | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| A | 0.1742 | 0.0746 | 0.1105 | 0.0609 | 0.0627 | 0.0826 | 0.0851 | 0.0568 | 0.0548 | 0.1102 | 0.0547 | 0.0555 |
| B | 0.0589 | 0.1995 | 0.1255 | 0.1161 | 0.0573 | 0.1876 | 0.056 | 0.2677 | 0.055 | 0.0582 | 0.055 | 0.0552 |
| C | 0.0593 | 0.0579 | 0.0578 | 0.0703 | 0.0557 | 0.2025 | 0.0556 | 0.0669 | 0.0611 | 0.0789 | 0.0543 | 0.0573 |
| D | 0.0587 | 0.0983 | 0.1659 | 0.1289 | 0.0583 | 0.1227 | 0.0566 | 0.1582 | 0.0581 | 0.0674 | 0.0563 | 0.0657 |
| E | 0.0572 | 0.1654 | 0.0576 | 0.0716 | 0.2404 | 0.0701 | 0.0605 | 0.0668 | 0.2287 | 0.0694 | 0.0607 | 0.0641 |
| F | 0.059 | 0.0595 | 0.0618 | 0.0624 | 0.2407 | 0.0595 | 0.0716 | 0.106 | 0.0816 | 0.0667 | 0.0593 | 0.0594 |
| G | 0.0562 | 0.129 | 0.0572 | 0.1867 | 0.1882 | 0.0841 | 0.0554 | 0.071 | 0.0572 | 0.0628 | 0.0751 | 0.0562 |
| H | 0.0589 | 0.1726 | 0.0578 | 0.0689 | 0.0575 | 0.0637 | 0.0578 | 0.0673 | 0.0607 | 0.0664 | 0.0586 | 0.2658 |
plate 08
| / | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| A | 0.0583 | 0.0592 | 0.0575 | 0.0929 | 0.0585 | 0.1161 | 0.232 | 0.119 | 0.0644 | 0.0592 | 0.0568 | 0.1757 |
| B | 0.0556 | 0.124 | 0.0803 | 0.0577 | 0.0546 | 0.071 | 0.0571 | 0.0702 | 0.0573 | 0.0599 | 0.0591 | 0.1539 |
| C | 0.0794 | 0.0621 | 0.056 | 0.1091 | 0.0678 | 0.0594 | 0.2024 | 0.0608 | 0.0676 | 0.061 | 0.0579 | 0.0778 |
| D | 0.0661 | 0.067 | 0.0584 | 0.0731 | 0.1041 | 0.1036 | 0.0695 | 0.0728 | 0.0735 | 0.0703 | 0.1152 | 0.1019 |
| E | 0.0659 | 0.2459 | 0.0572 | 0.0663 | 0.0555 | 0.1708 | 0.0579 | 0.0677 | 0.0577 | 0.0778 | 0.0623 | 0.1592 |
| F | 0.0606 | 0.0586 | 0.0587 | 0.0571 | 0.0721 | 0.227 | 0.0957 | 0.0634 | 0.1765 | 0.0702 | 0.0636 | 0.0607 |
| G | 0.054 | 0.0666 | 0.0564 | 0.0617 | 0.0567 | 0.1463 | 0.0576 | 0.0697 | 0.0564 | 0.2615 | 0.1307 | 0.06 |
| H | 0.1496 | 0.1021 | 0.0728 | 0.0785 | 0.0577 | 0.0726 | 0.0561 | 0.1611 | 0.0783 | 0.1801 | 0.0595 | 0.2327 |
First and Second Round Confirmation Screening: The 90 clones obtained from the initial screening were selected for further ELISA screening (with an additional 2 control clones). Detailed experimental data are shown in the table below.
Conclusion: After confirmation screening, 32 potential positive nanobodies were retained.
| / | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| A | 0.1263 | 0.0858 | 0.0863 | 0.0738 | 0.085 | 0.1102 | 0.0838 | 0.2779 | 0.2733 | 0.1157 | 0.0818 | 0.1336 |
| B | 0.1526 | 0.1888 | 0.0704 | 0.1205 | 0.0717 | 0.0801 | 0.0805 | 0.0833 | 0.0746 | 0.0753 | 0.2528 | 0.0771 |
| C | 0.0629 | 0.1208 | 0.0774 | 0.0876 | 0.2369 | 0.0768 | 0.1459 | 0.0728 | 0.114 | 0.0646 | 0.2313 | 0.0573 |
| D | 0.0796 | 0.113 | 0.0718 | 0.0755 | 0.1654 | 0.0958 | 0.0693 | 0.0693 | 0.0682 | 0.1419 | 0.0781 | 0.0584 |
| E | 0.0804 | 0.0692 | 0.0772 | 0.1529 | 0.068 | 0.0606 | 0.103 | 0.0683 | 0.0705 | 0.0798 | 0.1797 | 0.058 |
| F | 0.1057 | 0.1261 | 0.0785 | 0.0732 | 0.1378 | 0.0778 | 0.0995 | 0.1877 | 0.0657 | 0.3154 | 0.1249 | 0.0576 |
| G | 0.0908 | 0.079 | 0.0626 | 0.0795 | 0.1156 | 0.0841 | 0.0607 | 0.0888 | 0.0757 | 0.3253 | 0.1943 | 0.0612 |
| H | 0.0638 | 0.0943 | 0.094 | 0.0758 | 0.095 | 0.1342 | 0.2676 | 0.0871 | 0.093 | 0.0942 | 0.0728 | 0.2256 |
4 - Sanger Sequencing to Obtain Positive VHH Sequences
- The aforementioned nearly 32 clones were subjected to Sanger sequencing, resulting in a total of 32 valid VHH gene sequences.
- After translation and removal of errors and duplicates, a final set of 23 non-redundant VHH gene sequences was obtained.
- After recombinant expression and purification of the 23 monoclonal nanobodies, ELISA was conducted to assess the affinity between the nanobodies and IL18. Eventually, 19 effective nanobodies were obtained.