Research published in Molecular Therapy: Oncology by teams from the Institute for Integrative Biology of the Cell and the Curie Institute, among others, demonstrated the identification of VSV-G backbone optimization mutations (H22N and S422I) through experimental evolution, successfully overcoming the functional defects caused by N-terminal nanobody fusion. The researchers fused nanobodies targeting the HER2 receptor with the optimized VSV-G and introduced K47Q or R354Q mutations to eliminate LDL-R binding capability. The resulting chimeric glycoproteins enabled pseudotyped VSV (VSVΔG-GFP) and lentiviruses to specifically target and infect HER2-positive cells. This provides an efficient and safe technological platform for targeted oncolytic virotherapy and gene therapy of HER2-positive tumors, and lays the foundation for developing therapies against other targets.
Initial Chimera Construction and Backbone Optimization
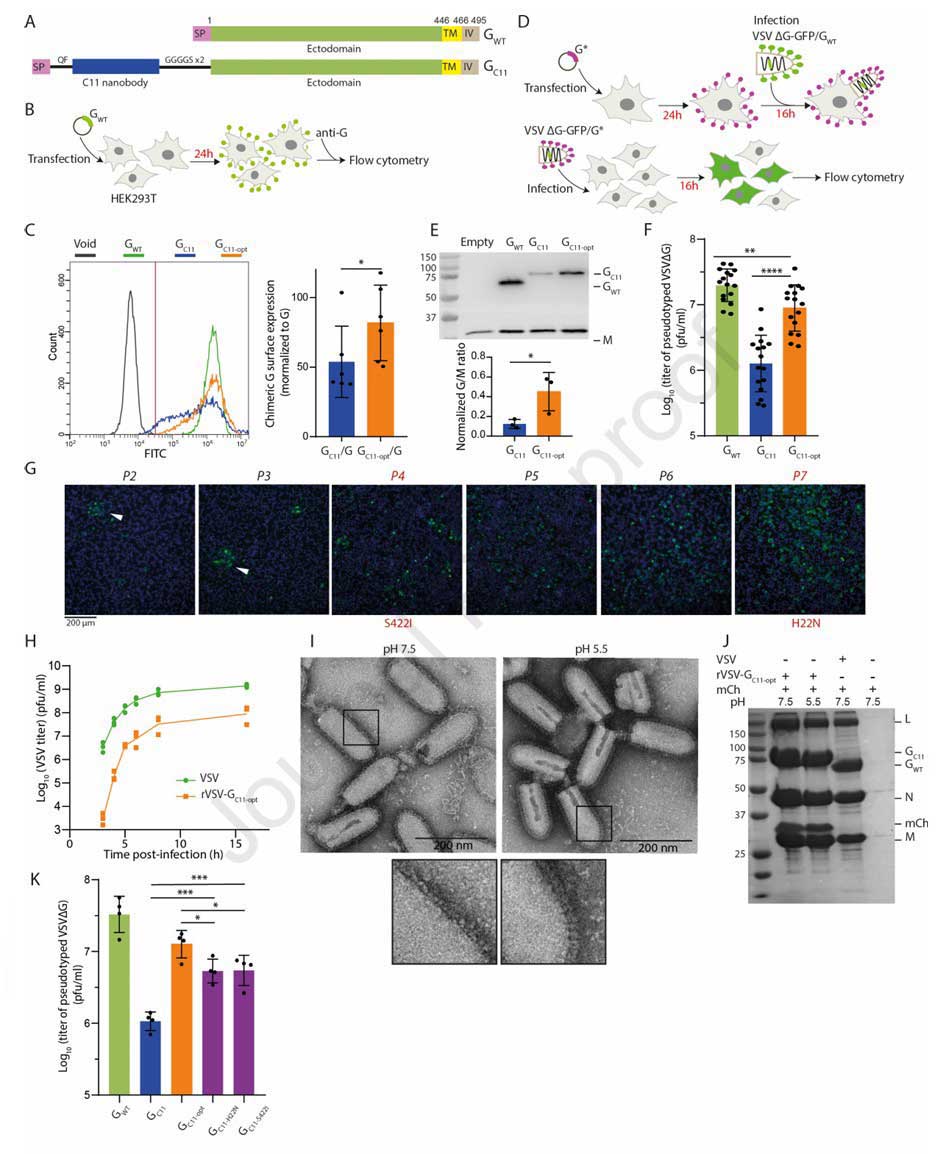
The research team first constructed a chimeric glycoprotein, GC11, targeting mCherry. They inserted the C11 nanobody, flanked by a QF dipeptide and a GGGGS×2 flexible linker, between the VSV-G signal peptide and its ectodomain. Transfection of HEK293T cells revealed that the cell surface expression level of GC11 was significantly reduced compared to wild-type VSV-G (VSV-GWT), reaching only 53 ± 23% of VSV-GWT.
Subsequent pseudotyping experiments further confirmed its functional impairment: the incorporation efficiency of GC11 into the VSVΔG-GFP envelope was lower than that of VSV-GWT, and the infectious titer of the pseudotyped virus was 1.3 Log₁₀ lower than that of VSV-GWT. This indicated that direct fusion of a nanobody to the N-terminus of VSV-G severely compromised its normal function.
To address this issue, the team employed experimental evolution: they replaced the G gene in the VSV genome with the GC11 gene to generate the recombinant virus rVSV-GC11, which was then serially passaged in BSR cells while monitoring viral titer and genetic mutations. Passaging results showed the emergence of the S422I mutation by passage 4, accompanied by a slight increase in viral titer. A second mutation, H22N, emerged by passage 7, forming a combination with S422I that led to a substantial increase in viral titer, surging from the initial ~10⁵ pfu/ml to ~10⁷ pfu/ml. By passage 10, the viral genome stabilized, containing only the H22N and S422I double mutations, with the endpoint titer at 16 hours post-infection stabilizing at ~10⁸ pfu/ml. This demonstrated the synergistic effect of the two mutations in restoring chimera functionality. This double mutant combination was designated GC11-opt and identified as the key optimized VSV-G backbone.
Figure 1: Core experiments and results of VSV-G backbone optimization
Functional Validation and Mechanistic Investigation of Optimized Mutations
To clarify the role of the double mutations, the team conducted multi-dimensional validation. Surface expression assays showed that GC11-opt expression on HEK293T cells reached 81 ± 24% of VSV-GWT, significantly higher than GC11. Western blot analysis indicated that the incorporation efficiency of GC11-opt into pseudotyped VSV particles was approximately 2-fold higher than that of GC11. In infection assays, the titer of VSVΔG-GFP pseudotyped with GC11-opt was 7-fold higher than that pseudotyped with GC11, approaching the level of VSV-GWT.
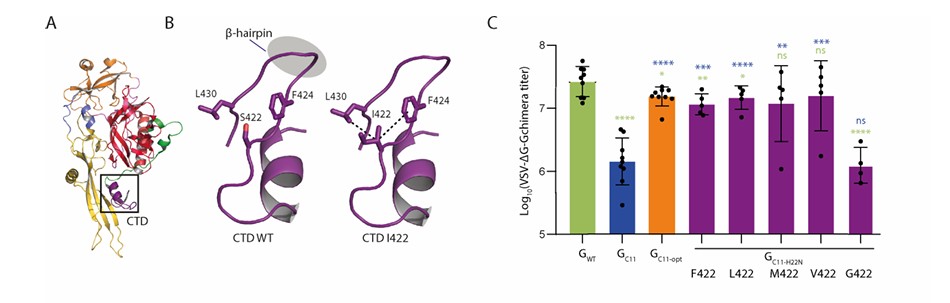
Further investigation into the S422 site revealed that replacing S422I with other hydrophobic amino acids resulted in pseudotyped virus titers not significantly different from GC11-opt. In contrast, replacement with the non-hydrophobic glycine reduced the titer back to the level of GC11. Structural analysis indicated that S422I stabilizes the β-hairpin structure of VSV-G through hydrophobic stacking interactions with neighboring residues F424 and L430. Although the molecular mechanism of H22N remains unclear, it significantly enhances the cell surface expression of the chimera. Together, these two mutations synergistically repaired the functional defects caused by nanobody fusion.
Figure 2: Effect of amino acid characteristics at VSV-G position 422 on nanobody fusion compatibility
Validation of Optimized Backbone Compatibility with Various Nanobodies
To confirm the general applicability of the optimized backbone, the research team fused 11 different nanobodies to both the original and optimized VSV-G backbones and compared the titers of pseudotyped VSVΔG-GFP.
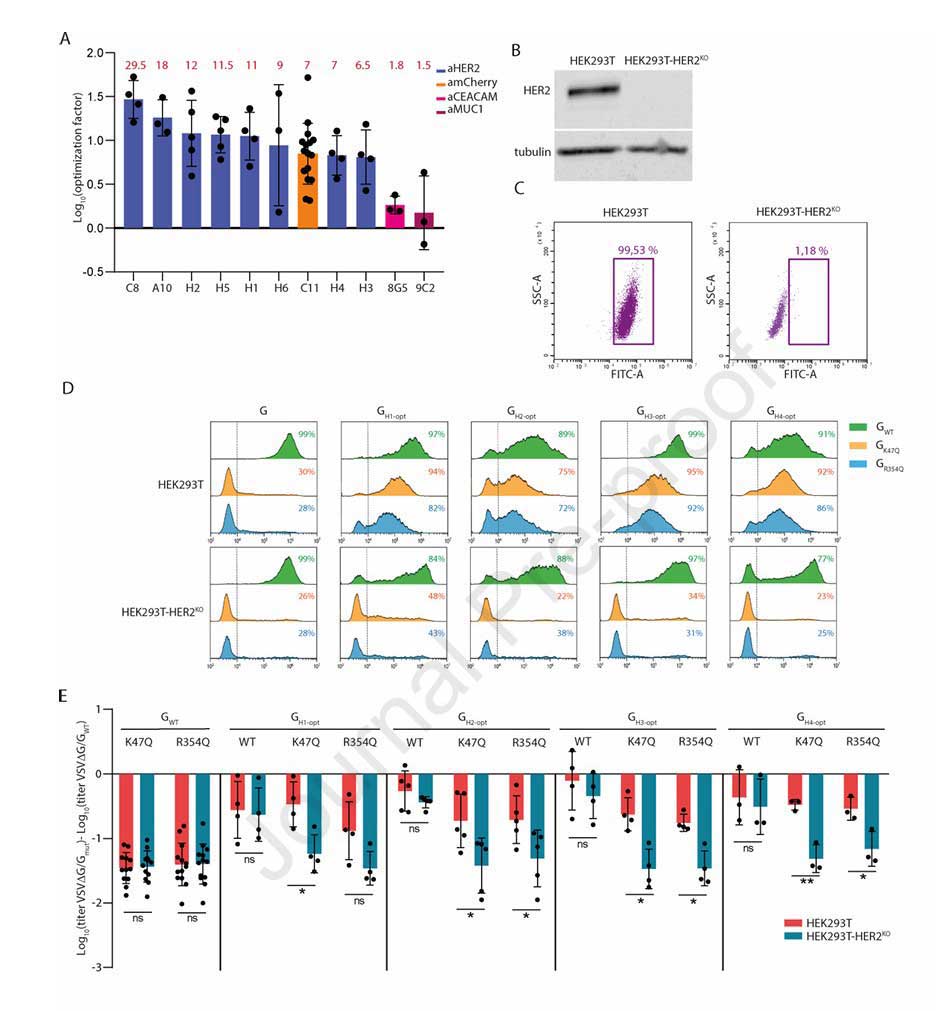
Results showed that for 9 out of the 11 nanobodies, the pseudotyped virus titer was significantly enhanced when using the optimized backbone, with enhancement factors ranging from 6.5 to 29 and a geometric mean of 11. The C8 nanobody showed the most pronounced optimization effect, with an enhancement factor of 29.5. The optimization factors for anti-HER2 nanobodies H1-H6 were all above 6.5, demonstrating the good compatibility of the optimized backbone with most nanobodies, particularly those targeting HER2.
Targeted Chimera Construction and Specificity Validation
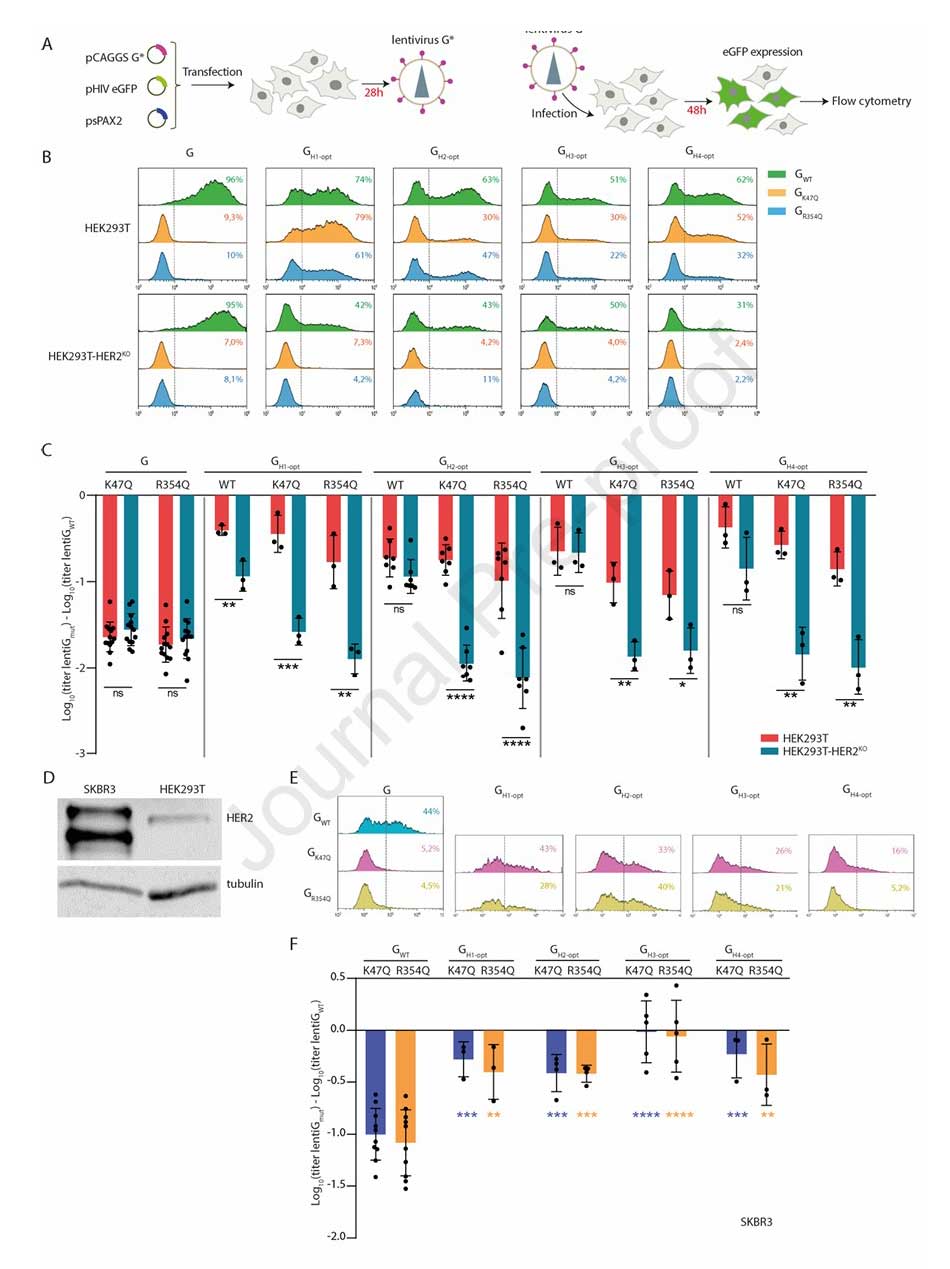
To achieve precise targeting of HER2-positive cells, the team introduced K47Q or R354Q mutations into the optimized HER2-targeting chimeras (e.g., GH1-opt, GH2-opt) to abolish LDL-R binding. Concurrently, they constructed a HER2 knockout cell line (HEK293T-HER2KO). Targeting specificity was validated by comparing infection efficiency in HEK293T (HER2-positive) versus HEK293T-HER2KO (HER2-negative) cells.
Figure 3: Functional optimization and targeting validation of anti-HER2 nanobody chimeras
Experimental results showed that in HEK293T cells, the titer of pseudotyped VSV carrying the anti-HER2 chimeras with K47Q/R354Q mutations was not significantly different from VSV-GWT. In contrast, their titer was significantly reduced in HEK293T-HER2KO cells, comparable to the non-targeting mutants containing only K47Q/R354Q (GK47Q, GR354Q). In SKBR3 breast cancer cells, which highly express HER2, the infection efficiency of pseudotyped lentiviruses carrying the anti-HER2 chimeras with K47Q/R354Q mutations was significantly enhanced by tens to hundreds of times compared to GK47Q/GR354Q.
Figure 4: Validation of infection specificity for HER2-targeting pseudotyped lentiviruses
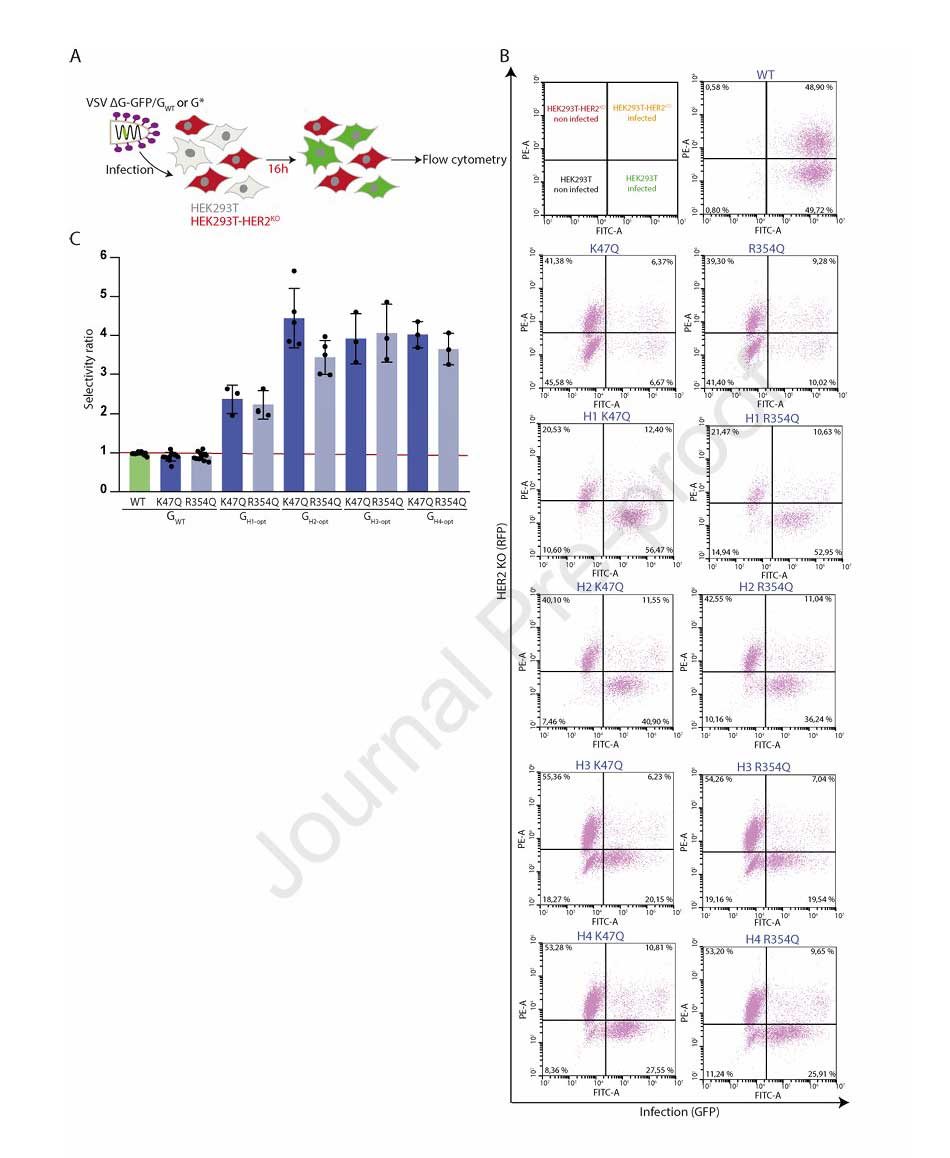
To further validate selectivity, the research team co-cultured HEK293T (HER2-positive) and HEK293T-HER2KO (RFP-labeled, HER2-negative) cells. They observed that pseudotyped VSV particles carrying the anti-HER2 chimeras incorporating K47Q or R354Q mutations demonstrated a selectivity index ranging from 2.25 to 4 for HER2-positive cells, conclusively demonstrating their specific recognition and infection capability towards HER2-positive cells.
Figure 5: Validation of targeting selectivity for VSV chimeras in mixed cell cultures
This study identified a highly versatile optimized VSV-G backbone that resolves the functional defects associated with N-terminal nanobody fusion. It enables precise targeting of VSV and lentiviruses to HER2-positive tumors, reduces off-target toxicity, and promotes the application of precise in vivo therapies. Furthermore, these chimeras can be incorporated into engineered extracellular vesicles for the targeted delivery of therapeutic cargoes such as drugs and genes, expanding the therapeutic landscape. The established "optimized VSV-G backbone + nanobody" technology platform demonstrates considerable generality, providing a novel strategy for developing new targeted therapeutics for various diseases.

Wuhan Nano Body Life Science and Technology Co. Ltd. (NBLST) is a nanobody industry platform established under the initiative of the Wuhan Industrial Innovation and Development Research Institute. Its headquarters is located in the main building of the Wuhan Industrial Innovation and Development Research Institute in the East Lake High-tech Development Zone, Wuhan. It boasts a 1400 m² independent laboratory in the Precision Medicine Industrial Base of Wuhan Biolake. Additionally, NBLST has established alpaca experimental and transfer bases in Zuoling, Wuhan, and Tuanfeng, Huanggang, both compliant with laboratory animal standards. These bases currently house over 600 alpacas, providing "zero-immunization-background" guaranteed alpaca immunization services for research institutions and antibody drug development companies.
NBLST focuses on the development, engineering, and application of nanobodies, and is dedicated to building an integrated public experimental service platform for production, education, and research. It possesses a full-chain technology platform encompassing antigen preparation (peptides, proteins, and RNA), antibody discovery and engineering, through to biological function validation/screening. The RNA antigens include RNA structurally and sequentially optimized for alpacas. Antibody discovery and engineering services employ multiple technological routes, including phage display, RNA, and mammalian cell display. Through cross-complementation of multiple platforms, it provides flexible antibody discovery and engineering services for pharmaceutical companies and research institutes, facilitating the development of drug reagents.
If you require our services, please feel free to contact us via email: marketingdept@nanobodylife.com
References:Duquénois, Isoline et al. “Optimization of the VSV-G backbone for amino terminal fusion with nanobodies allowing its specific retargeting to HER2 receptors.” Molecular therapy. Oncology vol. 33,4 201065. 25 Sep. 2025, doi:10.1016/j.omton.2025.201065